Computational engineering is an essential component in research that can benefit not only the academic and industrial world but also be the bridge that will cover the gap and bring collaborations that can move science and society many steps forward. Research in computational engineering are carried out in the following research themes:
- Multi-scale Computational Modelling of Nanostructured Materials with Applications in Nanotechnology
- Hybrid Physics-based Data-driven Approaches for Large-scale Simulations and AI Technologies
- Modelling of Biomolecular Systems for Biotechnology Applications
- Statistical Inference for Uncertainty Quantification and Model Selection
Multi-scale Computational Modelling of Nanostructured Materials with Applications in Nanotechnology
This direction considers nanostructured (complex fluids and macromolecular) materials, such as films and nanocomposites. Macromolecular systems (polymers) are characterized by the presence first, of chemical bonds joining the atoms into chains and, second, of intermolecular forces binding the macromolecular chains to one another. This feature (energy), together with the flexibility of the chains due to internal rotation of the units (entropy), and the interaction between different components-phases (interfaces) determines the properties of such hybrid systems.
Various technological issues are related to the coupling of interfaces with the macroscopic properties (mechanical, structural, dynamical, rheological, etc.) of such materials. For example, polymer and/or biopolymer/solid interfaces play a key role in applications involving polymer/particle nanocomposites, since the strength of chain attachment at interfaces (a quantitative thermodynamic measure of which is the work of adhesion) governs the mechanical properties of the structure. In addition, the organic/inorganic nature of their properties indicates their potential application as structural materials in renewable energy, drug design, aerospace, automotive, electronics and packaging industries.
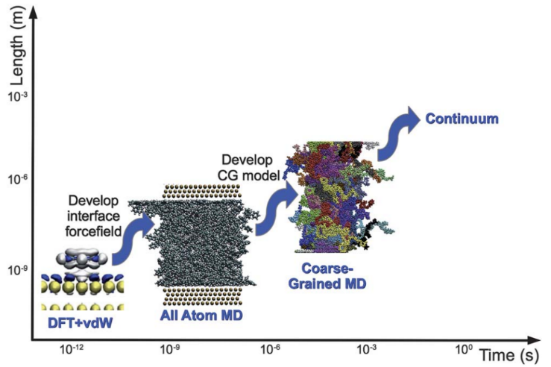
The main challenge in the computational study of the above systems is to predict their macroscopic properties directly from their molecular structure (structure-properties relations). This will allow the design of novel materials with the desired properties. To achieve this, multi-scale simulation methodologies that combine different levels of description are necessary, due to the enormous range of length and time scales characterizing such systems.
Based on our long time expertise we will apply such a hierarchical multi-scale methodology that combines different simulation techniques in a systematic way: From (a) ab-initio and DFT calculations of small systems, and (b) all-atom molecular dynamics, and Monte Carlo, simulations of large systems (of the order of 106 atoms), up to (c) rigorous mesoscopic coarse-grained (CG) models, and (d) continuum modelling and simulations, e.g. finite elements, that can be used to describe phenomena in the engineering scale (see Figure 2a). In all cases, results from the simulations at a more detailed scale are used to parametrize models at the less detailed (coarser) one.
The above hierarchical methodology, and the associated large-scale simulations involving millions of atoms/particles, will be used to predict structure-properties relations of several systems with important technological applications, such as:
- Graphene-based polymer nanostructured systems
- Polymer nanocomposites
- Thin films
- Systems under non-equilibrium conditions.
Important properties for the above systems will be predicted, including among others:
- Thermodynamics (pVT data, etc) and structure of the (fluid/solid, fluid/fluid or solid/solid) interface
- Mechanics (Young modulus, relaxation modulus)
- Dynamics and rheology of complex fluids (e.g. relaxation times, diffusion coefficient, viscosity, friction coefficient)
- Wetting properties.
Hybrid Physics-based Data-driven Approaches for Large-scale Simulations and AI Technologies
The main scientific topic of this direction is to use, and develop new as well, multi-scale computational (HPC + data-driven) methodologies in order to construct more accurate quantitative models of complex materials across different scales. To achieve this a hybrid physics-based data-driven paradigm is proposed, that links together high-performance computing (large-scale simulations) and AI technologies. In more detail, we envisioned work along the following directions.
Data-driven coarse-graining strategies
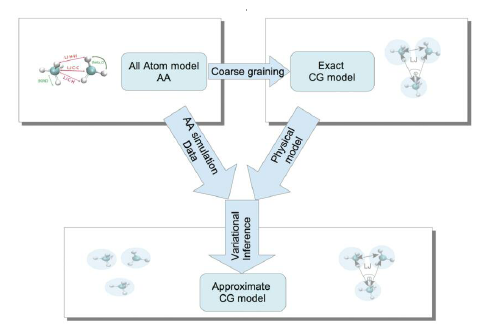
The development of data-driven systematic CG techniques is a very important and still unexplored area of multi-scale modelling. In systematic “bottom-up” strategies the effective CG interactions are derived as follows: Assume a microscopic system, composed of N atoms/particles in the canonical ensemble, described by a Hamiltonian HN in the microscopic q (3N) configuration (positions of all atoms), and a mesoscopic CG description of this system with M “superatoms” (M<N), and Hamiltonian HM, in the mesoscopic Q (3M) phase space.
The observable can be distribution functions (e.g. bonded distributions, pair correlation functions, g(r)), the total force acting on a CG particle, or the relative entropy, or Kullback-Leibler divergence, between the microscopic and the coarse space Gibbs measure. Despite the success of such methods, they become problematic for multi-component nanostructured systems (e.g. blends, interfaces, crystals, etc.) due to the complex heterogeneous structure, at the atomic level, of such systems. One of the reasons the above techniques fail is that the set of basis functions used to approximate the exact, but not computable, many-body PMF is not large enough.
To overcome the above limitations here we combine the existing schemes with deep learning-based approaches to provide more accurate, and more transferable as well, approximations of the CG model (free energy surface, FES) under a broad range of conditions. For this, we propose to use neural networks, NNs, which can, in principle, be used to fit any continuous function on compact subsets of Rn (see universal approximation theorem). However, their application to describe the interaction between atoms or molecular is not straightforward, since NNs do not obey the required symmetries (such as permutation, translation, etc) followed by physical laws in nature. Thus here the NNs will be combined with proper transformations of the original data. We anticipate that new CG force fields will be a more accurate approximation of the “exact” many-body PMF thus deriving in more powerful and transferable CG models. In addition, we will explore CG density-dependent potentials in which density-dependent terms are used to approximate the exact many-body PMF in an analogous manner to classical density functional theory. Recently we’ve developed such CG models for homopolymer bulk systems. Here we will extend such potentials to complex nanostructured systems, also comparing against the data-driven (NNs-based) approximations.
Linking microscopic and mesoscopic scales
The main challenge in the multi-scale modelling of complex materials is the systematic linking of the models across the different scales. For these algorithms that either eliminate (dimensionality reduction) or re-introduce degrees of freedom (back-mapping process) are required. Recently, we’ve developed hierarchical back-mapping strategies incorporating generic different scales of description from blob-based models and moderate coarse-grained up to all-atom models (see Figure 3). The central idea is to efficiently equilibrate CG polymers and then to re-insert atomistic degrees of freedom via geometric and Monte Carlo approaches. Furthermore, more recently we introduce a general image-based approach for structural back-mapping from coarse-grained to atomistic models using adversarial neural networks.
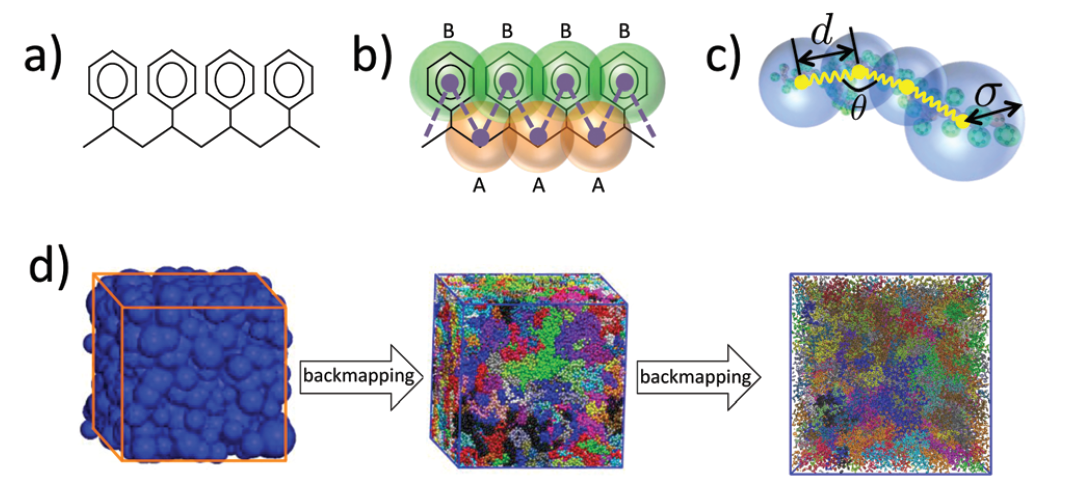
The above methods have been extensively tested for polymer melts of high molecular weight. Here, we will extend these methods to provide large all-atom configurations for heterogeneous nanostructured systems. This is a particularly challenging area, due to the inherent complexities of such systems, that has not been addressed in the literature so far. The new methods will be thoroughly examined and validated by comparing their structural and conformational properties of the back-mapped model configurations with reference data from smaller systems, which are obtained directly from long atomistic simulations.
Modelling of Biomolecular Systems for Biotechnology Applications
The third direction of the proposed research program concerns the study of biomolecular systems with important applications in biotechnology, which is nowadays a very active and intense research field. Research is carried out on the following fields.
Bio-based films and surfaces
The development of protein-based films and coatings with the desired properties can have strong technological impact within the circular economy era. For example, such films could be used to replace existing synthetic oxygen barrier polymers and increase the recyclability of the packaging products. Thus, the study of functionalized biodegradable monolayer films with different functional (barrier, mechanical, optical, etc.) and active properties at the molecular level is a very challenging research field.
The prediction of structure-properties relations of bio-based complex materials is directly connected to their study at the molecular level. In this aspect, a synergy of multi-scale simulation techniques and AI technologies could be an important factor for the design of functional bio-based materials. Our main goal is to pursue such a fundamental (molecular-level) understanding of structure-processing-functionalization-property relationships in bio-based materials via large-scale simulation methodologies and machine learning methods.
We will apply molecular simulation tools such as molecular dynamics (MD) and Monte Carlo (MC) to get a microscopic understanding of structural, conformational and dynamical properties of biomolecules (peptides, proteins, carbohydrates, lipids) in different environments, substrates and conditions. We aim to use these results in order to provide: (a) a detailed characterisation of functionalized bio-based interfaces for targeted applications, and (b) a molecular understanding of properties of bio-based thin films, and guide their design.
Abiotic-biotic interactions of bio-derived surfaces and their environment
Furthermore, we plan to use the above-described methodology for the development of materials with anti-bacterial properties. For this, we will study interactions, at the molecular level, between nanostructured materials (inorganic and bio-based) and peptides, and quantify the strength of binding of peptides to specific minerals/ materials. Our goal is the identification of the structural features in the peptides and of surface chemistry on biomolecule binding that lead to enhanced antibacterial behaviour, and on the long-term to develop rules to predict binding behaviour for different materials.
Due to the high complexity of the effect of surface chemistry and topology on biomolecule binding we propose: (a) many large-scale simulations of specific systems, and (b) Data analytics approaches for analysing simulation, and experimental data. Molecular simulations will be used in both atomic-resolution and simplified coarse-grained (CG) representations to study the influence of amino acid sequence and functionality on the binding process of the peptides of interest onto selected surfaces.
Examples of expected results from the above methodology include. 1) Molecular-level understanding of the effect of important environmental parameters (solvent, temperature, pH) and structural factors (composition, chain length, backbone flexibility, etc.) on peptide or protein self-organization, microstructure, adhesion, packing on substrate, and antimicrobial activity. 2) Better understanding of cooperative effects during binding (such as configurational re-arrangements followed by strong adsorption on the substrate), and of the equilibrium state after binding. 3) Based on the above, alongside with peptide conformation data, propose design rules for optimized coating-substrate interactions, having also the desired antimicrobial activity, for specific applications.
Nanocomposites of biomolecules and nanoparticles (NPs)
We also plan to extend the above methods to examine interactions of biomolecules with inorganic nanoparticles. Our goal is to provide quantitative information for the structure of organic molecules and proteins through multi-scale dynamic simulation approaches, involving detailed all-atom models and proper coarse-graining strategies (as described in Theme 2).
We will predict structure, conformations, and dynamics of biolpolymers/NPs model systems, such as PLA/Silica, as a function of polymer/NP interaction, temperature, concentration etc. In addition, the above computational scheme will allow us to: 1) Perform large-scale all-atom MD simulations to study proteins/nanoparticles interface at atomistic scale, 2) Develop effective potentials for complex protein structures for use in coarse-grained (CG) simulations of these systems, to get in a more efficient way simulation predictions for the relevant properties and 3) Provide quantitative predictions about microstructure and dynamics of biomolecules interacting with nanoparticles.
Statistical Inference for Uncertainty Quantification and Model Selection
Uncertainty Quantification (UQ) is the process of measuring the uncertainty in the parameters of a physical model that are calibrated on experimental observations. Statistical (e.g. Bayesian) inference is one of the standard techniques for UQ that incorporates both expert knowledge (prior) and experimental measurements (likelihood). The uncertainty in the parameters can be propagated to any quantity of interest achieving updated robust predictions. In addition, UQ offers the model evidence, a measure of fitness of the model to the data.
Our plan is to develop efficiently parallelized optimization and sampling algorithms casted in terms of Bayesian UQ. Other ML methodologies, related also to the field of molecular modelling by trying to augment the classical physics-based potentials, will be implemented, using supervised and/or unsupervised learning, such as the Neural Networks, Gaussian process regression, SVM, etc.
The data-driven variational inference will be described and implemented via a “Digital Twin” paradigm, shown below. In more detail, we propose work along the following areas.
Data-driven Confidence interval estimation in coarse-graining
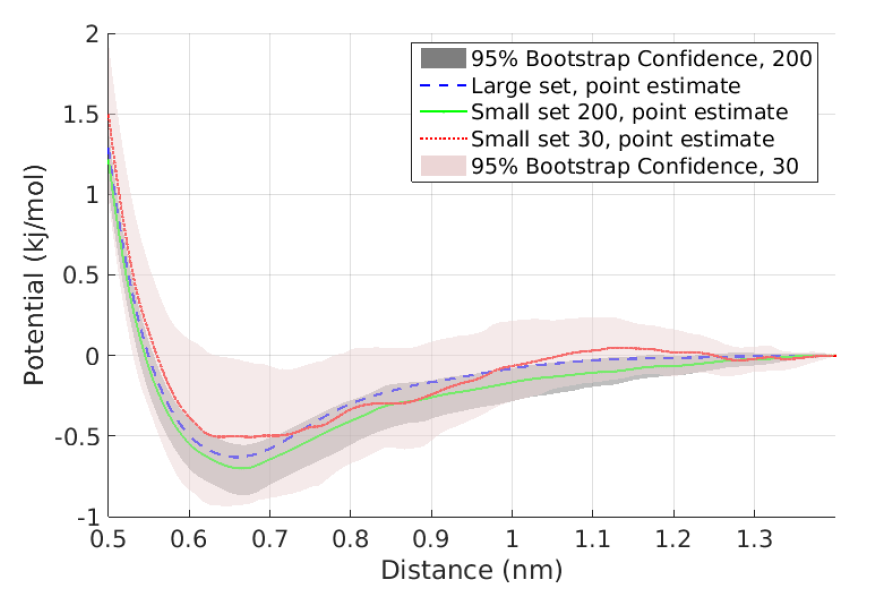
The quantification of induced errors, in bottom-up systematic CG models, due to the limited availability of fine-grained data is still an unexplored area of mathematical CG. Here we propose new methodologies and algorithms for the quantification of confidence in bottom-up coarse-grained models for molecular and macromolecular systems, based on rigorous statistical methods. The proposed method is based on a recently developed statistical framework by us in which statistical approaches, such as bootstrap and jackknife, are applied to infer confidence sets for a limited number of samples, i.e. molecular configurations. Moreover, we gave asymptotic confidence intervals for the CG effective interaction assuming adequate sampling of the phase space.
These approaches have been demonstrated, for both independent and time-series data, in a simple two-scale fast/slow diffusion process projected on the slow process. They have also applied on an atomistic polyethylene (PE) melt as the prototype system for developing coarse-graining tools for macromolecular systems. For this system, we estimate the coarse-grained force field and present confidence levels with respect to the number of available microscopic data. The above approaches will be extended to deal with more challenging multi-component nanostructured heterogeneous materials, as those described in Themes 1 and 3, and also for systems under non-equilibrium conditions (eg. shear flow).
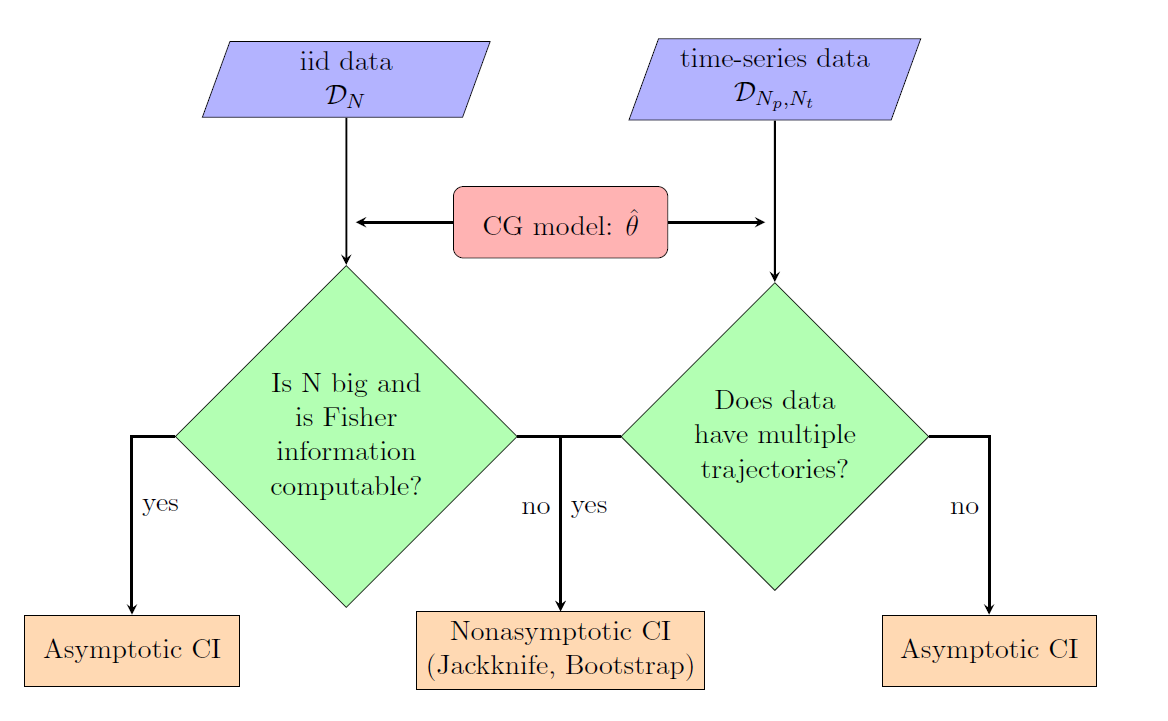
Force field Selection and Sensitivity Analysis of model parameters via information-theory approaches
Furthermore, we will derive a systematic parametric sensitivity analysis (SA) methodology for the CG effective interaction potentials derived n the framework of the current project. Our SA method is based on the computation of the information-theoretic (and thermodynamic) quantity of relative entropy rate (RER) and the associated Fisher information matrix (FIM) between path distributions, that has been applied before by us on two different molecular stochastic systems, a standard Lennard-Jones fluid and an all-atom methane liquid. The obtained parameter sensitivities were compared against sensitivities on three popular and well-studied observable functions, namely, the radial distribution function, the mean squared displacement, and the pressure. Here, we will extend this approach for multi-component nanostructure systems.
In addition, the above methods will be combined with the Hierarchical Bayesian framework to be used as a selection tool for (a) force field selection in MD simulations using heterogeneous data and (b) for the selection of the appropriate coarse-grained model for a given system.
Related Projects
 |
|
|
|
|
People